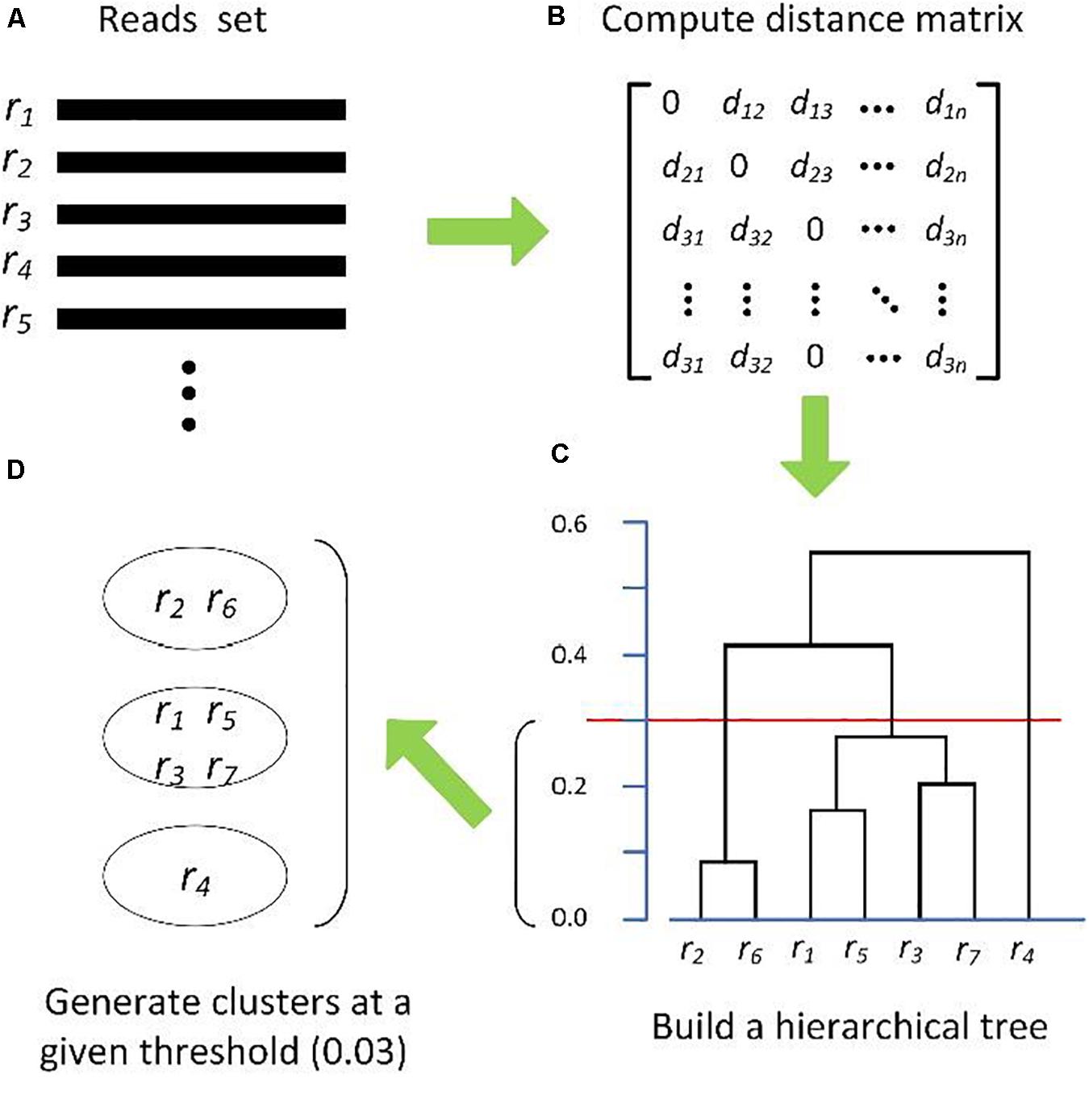
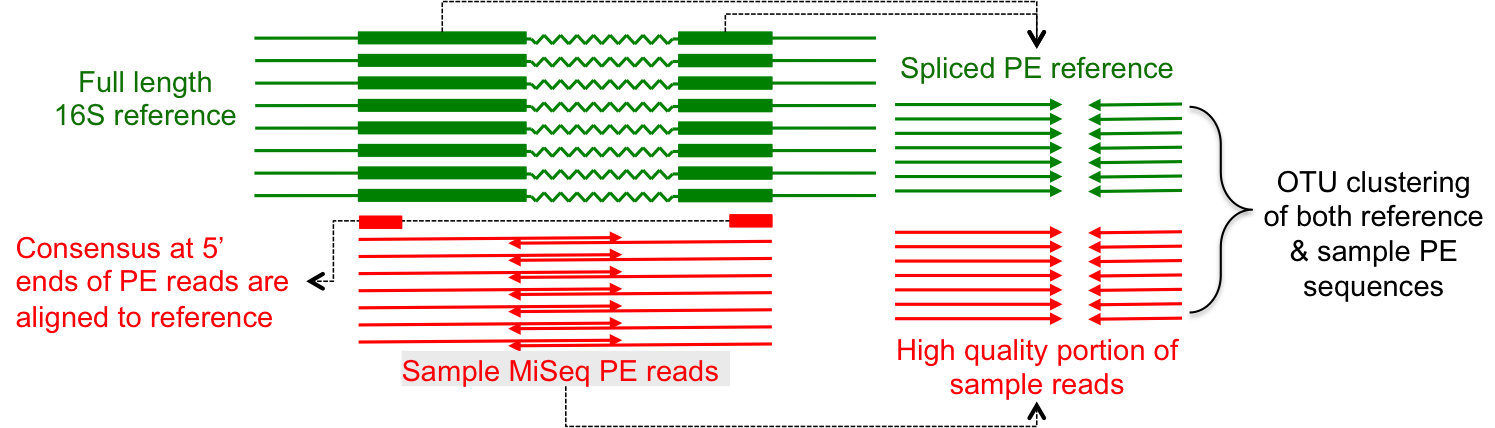
Clustering biological sequences with dynamic sequence similarity threshold | BMC Bioinformatics | Full Text

Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences | SpringerLink
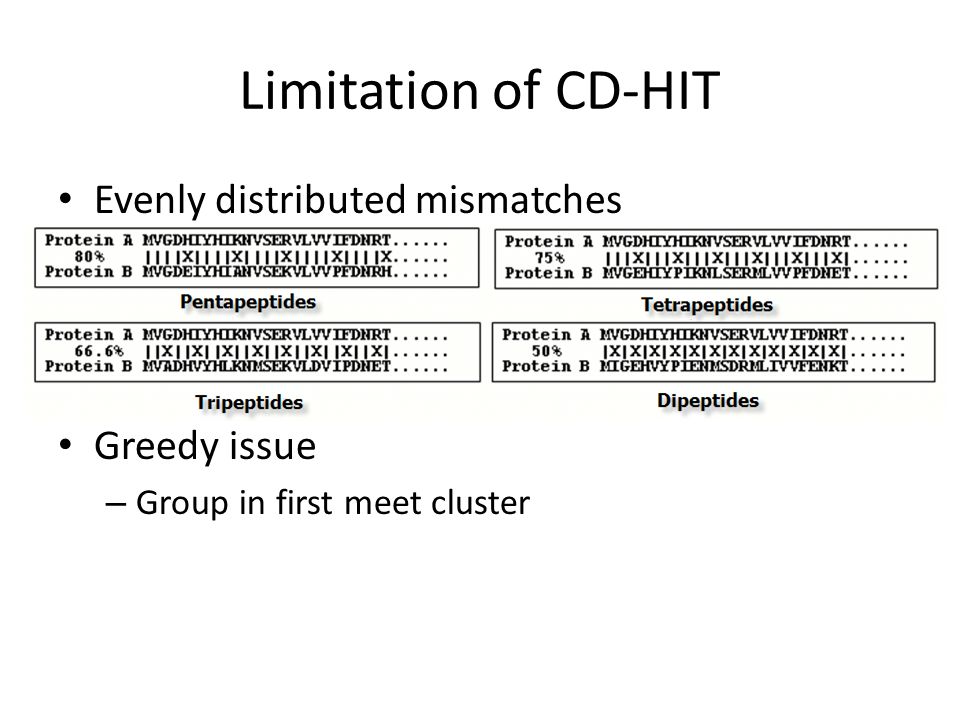
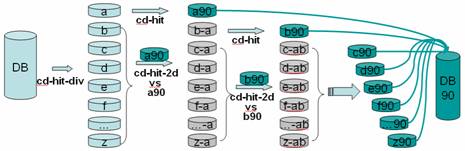
Analysis and comparison of very large metagenomes with fast clustering and functional annotation Weizhong Li, BMC Bioinformatics 2009 Present by Chuan-Yih. - ppt download

An example illustrating how the CD-HIT main paradigm works. Record 1 is... | Download Scientific Diagram

kClust: fast and sensitive clustering of large protein sequence databases | BMC Bioinformatics | Full Text
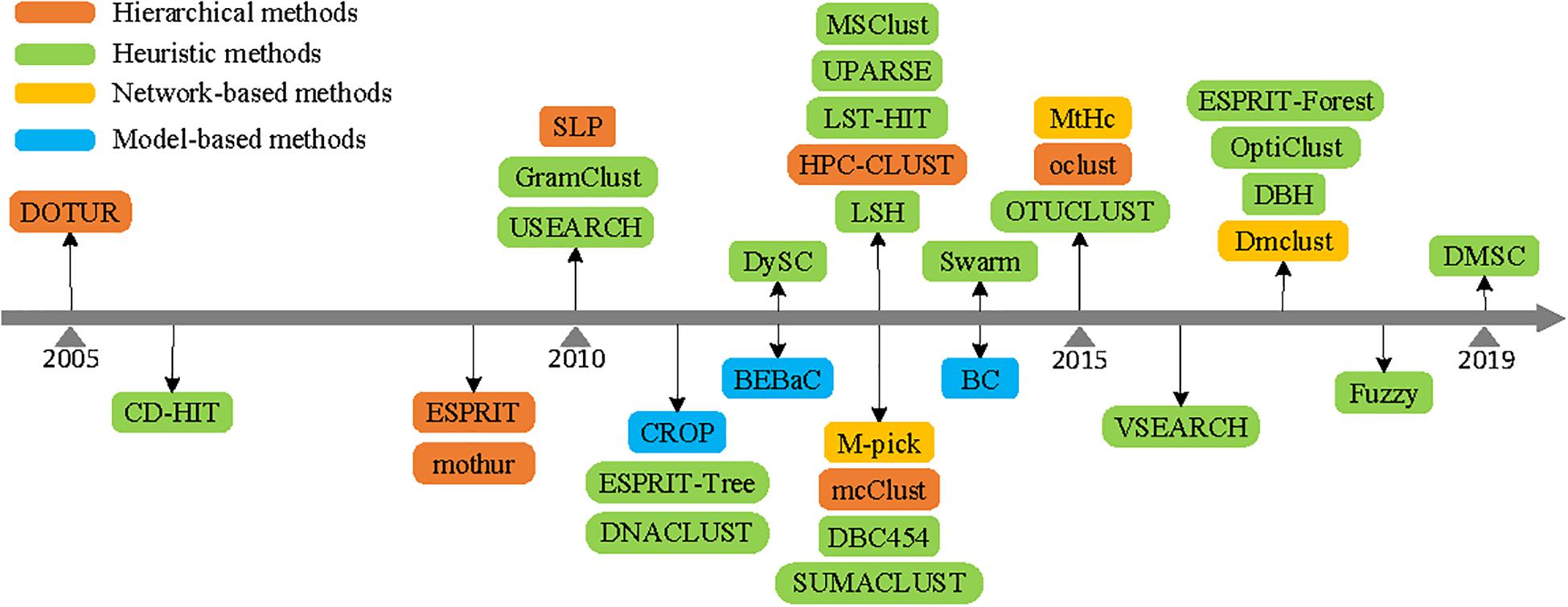
Frontiers | Comparison of Methods for Picking the Operational Taxonomic Units From Amplicon Sequences

An example illustrating how the CD-HIT main paradigm works. Record 1 is... | Download Scientific Diagram

A comparison of the performance of different clustering approaches. For... | Download Scientific Diagram
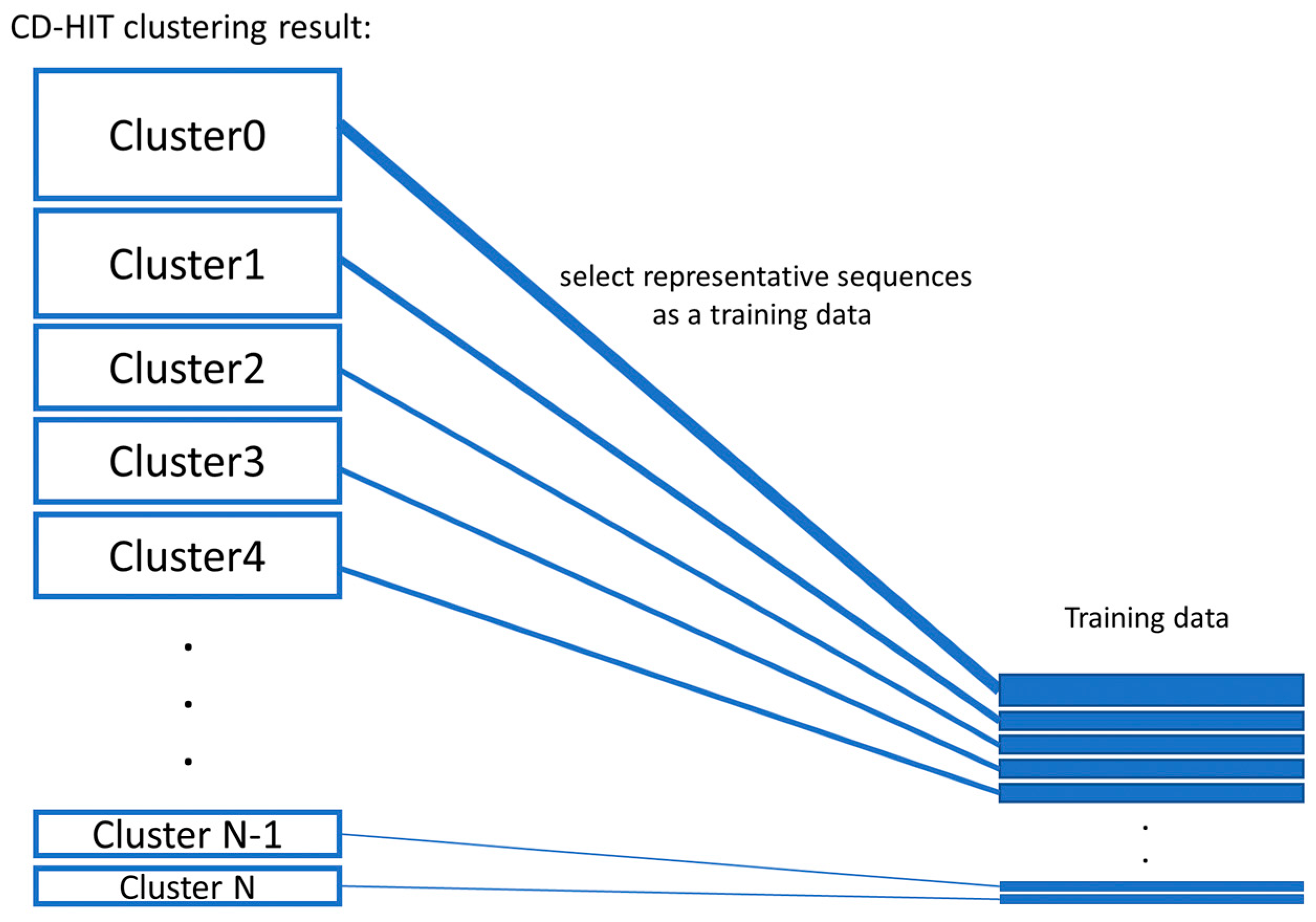
Genes | Free Full-Text | Ensemble-AMPPred: Robust AMP Prediction and Recognition Using the Ensemble Learning Method with a New Hybrid Feature for Differentiating AMPs
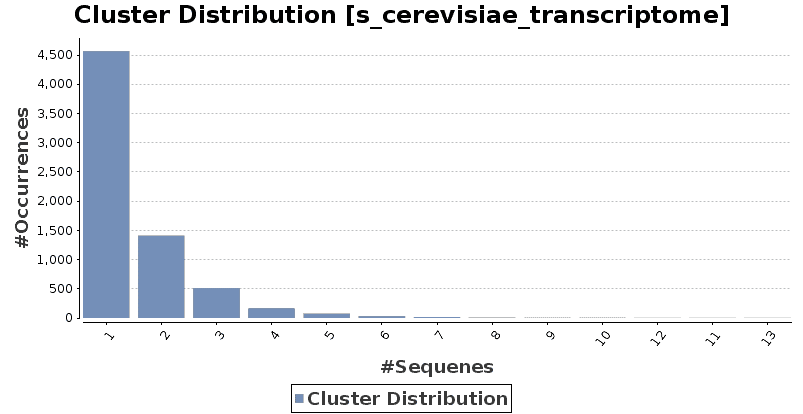
De Novo Assembly of the Transcriptome of the Non-Model Plant Streptocarpus rexii Employing a Novel Heuristic to Recover Locus-Specific Transcript Clusters | PLOS ONE

Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences | SpringerLink